
Clinical Research Analytics Environment (CRANE)

What is the Clinical Research Analytics Environment at UNMC?
UNMC’s Clinical Research Analytics Environment (CRANE) is a flexible, inclusive clinical data warehouse combined with the tools, processes, and people needed to support knowledge discovery. It contains de-identified, structured data for UNMC research and quality improvement efforts. CRANE can be used as a search discovery tool allowing qualified researchers to search de-identified patient data from sources that would otherwise require IRB-approval or asynchronous consideration. CRANE is considered an environment because it is more than just the software and available data. CRANE is an entire ecosystem consisting of an IRB-approved, de-identified clinical data warehouse (CDW) that provides researchers with access to well organized, characterized, and standardized patient-level data in compliance with HIPAA, the common rule, and best practices. This CDW consolidates data from a variety of clinical sources to present a singular, multi-faceted view of the available data.
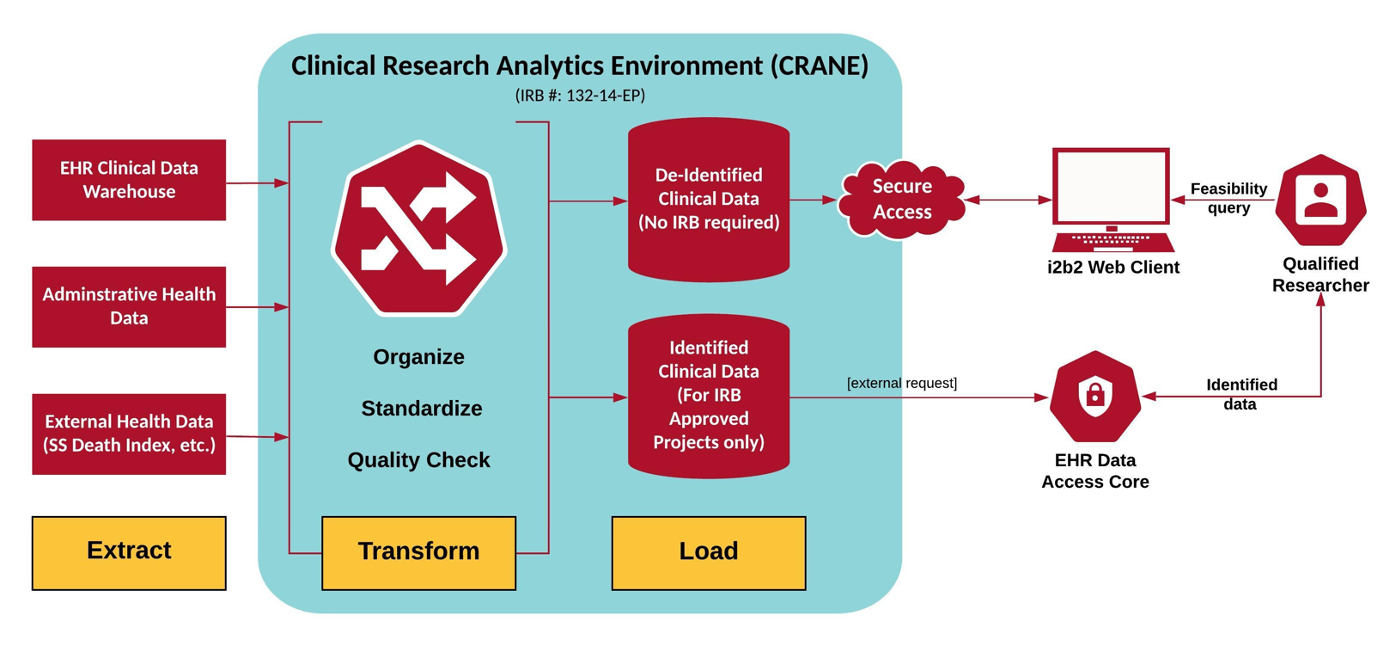
The primary data in CRANE derives from the Nebraska Medicine Electronic Health Record (EHR) system through a series of extracts and transformations to render the data in a well-characterized, interoperable format based on the PCORnet Common Data Model. The EHR data is supplemented with other patient-level data sources including, but not limited to, state cancer registry data, encounter data from the Nebraska Health Information Initiative, and the Social Security death index. Additional data sources to expand the patient-level data include formally encoded anatomic pathology data, biomarker data, and pointers to biobank specimens. This creates a rich repository for health research as it is actually happening in a clinical setting.
The data in CRANE is rendered in the Office of the National Coordinator for Health Information Technology (ONC) approved standard codes and supports external analysis by both R and SAS statistical software. The de-identification process involves comprehensive removal of all 18 PHI identifiers defined by HIPAA as well as a complex date shifting process. Date shifting is inconsistent among patients but is consistent at the individual patient level for each instance of CRANE. This results in dates that are shifted between 1 and 30 days in the past across all of a patient’s data points.
In an effort to support clinical researchers without exposing them to the technical details, CRANE uses an integrated approach providing a “self-serve” data mechanism for qualified researchers. Researchers interact with the data in CRANE primarily through the i2b2 (Informatics for Integrating Biology with the Bedside) web client. I2b2 is a cohort discovery tool developed by Harvard informaticists that allow researchers to determine if there is a cohort of patients in our clinical data repository that meets their criteria of interest using a drag-and-drop interface for ease of use. In summary, CRANE is the entire research environment while i2b2 is the web client portion of that environment through which researchers engage with the underlying data.
To submit a data request click here
A sample of some of the research derived from CRANE is available on our News, Research, and Development Projects page.
What is a qualified researcher?
Access to the system is limited to UNMC faculty or their designees who have completed CITI training and are in compliance with UNMC competency training, which includes HIPAA and Information Security Awareness training. Access to identified patient data requires IRB approval. The data release is through an honest broker for qualified faculty.
What does CRANE represent?
The research data marts exposed for use represent a detailed distillation of the raw EHR data normalized, standardized and processed with supporting metadata to assist researchers in calculating computable phenotypes – which is a clinical condition, characteristic, or set of clinical features determined exclusively from the data available in EHRs and ancillary data sources without chart review or interpretive analysis by a clinician – for clinical research. Within CRANE are a number of data marts organized to meet the needs of collaborating Research Networks.
The National Patient-Centered Clinical Research Network publishes a detailed Common Data Model (CDM) that is linked to a secure mechanism for querying. The current version of the CDM is v.5.1, which was released in September 2019. PCORnet writes SAS code to query the CDM for supported trials. Because these are national trials with 50-80 data nodes, there is rigorous data quality checking and data characterization.
The Greater Plains Collaborative (GPC) is the prime mover behind our local development of CRANE. The 12 GPC member academic medical centers maintain architecturally similar systems allowing collaborative development of the technology. The GPC supports a query mechanism where local queries are shared through the query compiler BABEL. These queries require local customization, so each site then customizes the query to work locally.
In order to further improve query sharing, the GPC is collaborating with Harvard’s clinical informatics team to deploy a large-scale flexible data mart within i2b2 termed Scalable Collaborative Infrastructure for a Learning Healthcare System (SCILHS). This allows shared queries through the Harvard SHRINE networked query tool. The SHRINE system operates through secure channels to individual data marts.
CRANE also provides a powerful clinical informatics education and research platform for extending data standardization and linking. A Big-Data-to-Knowledge U01 grant supports the process of linking anatomic pathology findings, biomarkers, and biobank data with the EHR data in CRANE. The resultant architecture is under development internationally, led by UNMC informatics researchers.
CRANE Connections – PCORnet and PopMedNet
- UNMC in partnership with Great Plains Collaborative (GPC) Network (11 other Midwestern academic medical institutes) received funding from the Patient Centered Outcome Research Institute (PCORI) to implement I2B2 & PopMedNet.
- The I2B2 backend connects to the Integrated Clinical Research Data warehouse (ICRD) of UNMC.
- Currently CRANE is operational from the de-identified version of ICRD in support of PCORnet sponsored Comparative Effectiveness Research Trials.
- PopMedNet is software developed by Harvard Medical School. UNMC’s Research IT Office built the production environment for PopMedNet.
- The PopMedNet software application enables simple creation, operation, and governance of distributed health data networks. It facilitates distributed analyses of electronic health data to support medical product safety, comparative effectiveness, quality, medical resource use, cost-effectiveness, and related studies.
- The PopMedNet enables UNMC to securely receive and send query data held by GPC partners. The software also allows GPC partners to maintain physical and operational control over their data.
In summary, the I2B2 software framework connects to the de-identified version of ICRD and can be used by UNMC & Nebraska Medicine researchers. PopMedNet also connects to the de-identified version of ICRD but serves GPC network partners.
CRANE Training Opportunities
In an effort to support CRANE at the departmental level, the CRANE team has been training superusers across campus. These users are experts at navigating the system and are more experienced in specific Epic modules. The initial class of Superusers graduated from the course in June 2019. These superusers form the foundation for data access and assistance.
The next round of Superuser training is scheduled for the first 8-week session in Summer 2020 (May 18 – July 10). It will run on Wednesday from 4:00 – 6:00 PM. If you’re interested in becoming a Superuser or would like to see if your department has a Superuser, please contact Jerrod Anzalone.
Additionally, self-learning tools are in development for a wide audience and are expected to be available in Summer 2020. This will be designed for users of all levels. Stay tuned.
Citing CRANE in Publications
Please use the following language to cite CRANE in your publication, presentation, press release, or RFA:
The project described utilizes the UNMC Clinical Research Analytics Environment (CRANE). CRANE is supported funding from the National Institute of General Medical Sciences, U54 GM115458 and the Patient Centered Outcomes Research Institute, PCORI CDRN-1306-04631. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or PCORI.
